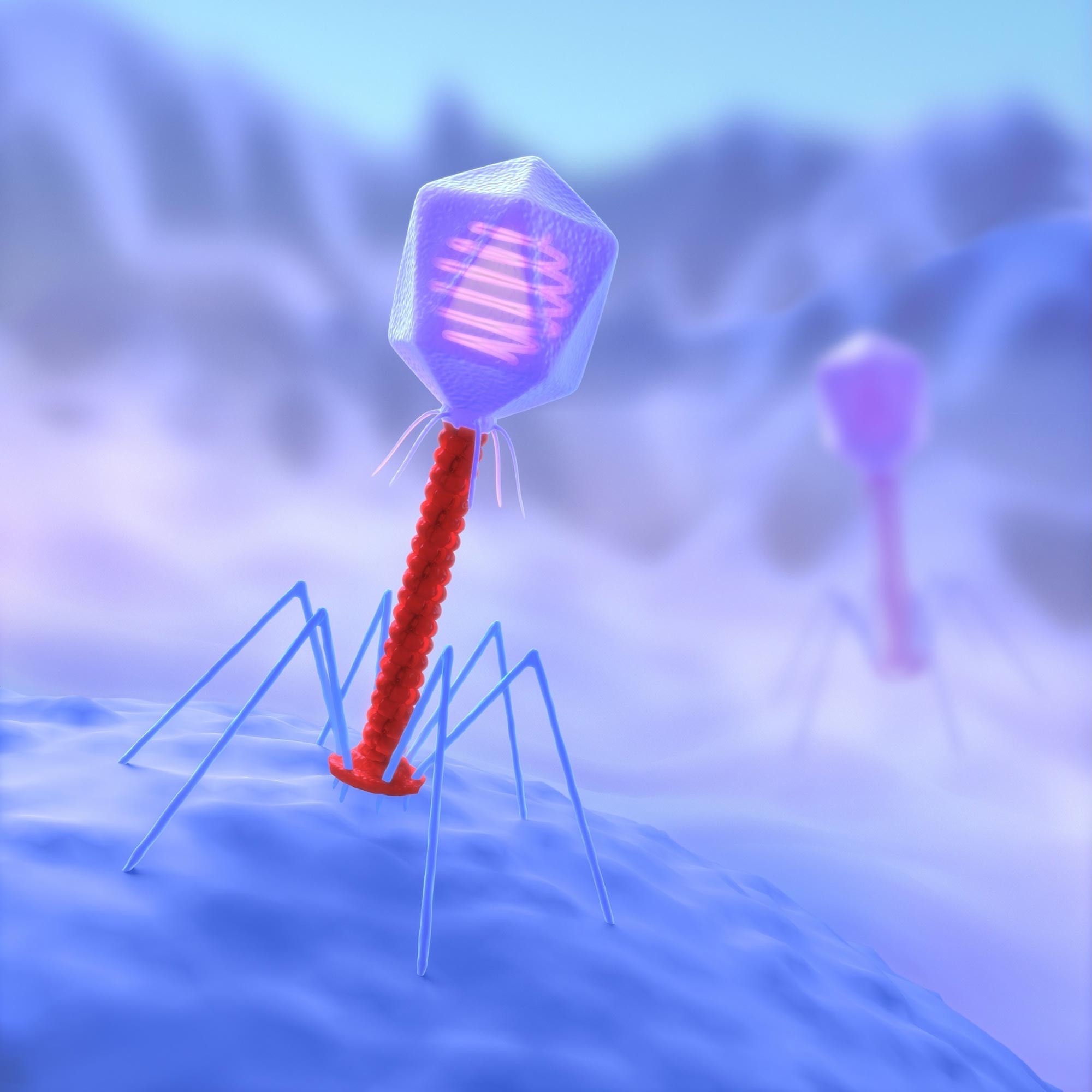
Phage illustration. Tailocins look like phage, but do not have the cap (‘head’) that contains the viral DNA and replication machinery.
A Berkeley Lab-led team is investigating the bizarre bacterial-producing nanomachines that could advance microbiome science.
Imagine there are arrows that are deadly when fired at your enemies, yet harmless when dropped on your friends. It’s easy to see how it would be a great advantage in warfare if it were real. However, something like these arrows does exist, and it is used in warfare … only on a different scale.
These weapons are called tailocins, and reality is almost stranger than fiction.
“Tailocins are extremely powerful protein-nanomachines produced by bacteria,” said Vivek Mutalik, a researcher at Lawrence Berkeley National Laboratory (Berkeley Lab), who studies tailocins and phage, the bacterial-infecting viruses that tailocins look like, are remnants. explain. ‘They look like phage, but they do not have the cape, it is the’ head ‘of the phage that contains the viral. DNA and replication machinery. It is thus like a needle driven on the spring that sits on the target cell, and then it appears to pierce the cell membrane and make a hole in the cytoplasm, causing the cell to lose its ions and contents and collapse. shower. ‘

An illustration of tailocins, and their altruistic action painted by the author Vivek Mutalik’s daughter, Antara. Credit: Antara Mutalik
A large variety of bacteria can produce tailocins, and they appear to do so under stress conditions. Since the tailocins are only lethal to specific strains – so specific that they have earned the nickname “bacterial homing missiles”, it appears that tailocins are a tool used by bacteria to compete with their competitors. Because of their similarity to phage, scientists believe that the tailocins are produced by DNA that was originally inserted into bacterial genomes during viral infections (viruses instruct their hosts to make more of themselves), and over evolutionary time the bacteria have the parts of the phage DNA that was not beneficial but retained the parts that could be co-opted for their own benefit.
But unlike most abilities selected by evolution, tailocins do not save the individual. According to Mutalik, bacteria are killed when they produce tailocins, just like when they are infected by true phage virus, because the pointed nanomachines burst through the membrane to leave the producing cell, just like repeated viral particles. But once the tailocins are released, they are only targeted at certain strains, saving the other cells of the host line.
‘They favor family members, but the individual is sacrificed, which is a kind of altruistic behavior. But we still do not understand how this phenomenon happens in nature, ”Mutalik said. Scientists also do not know exactly how the needle of the tailocin works.
These topics, and tailocins as a whole, are a research area because of the many possible applications. Mutalik and his colleagues at Berkeley Lab’s Biosciences Area, along with collaborators from UC Berkeley, would like to use tailocins to better study microbiomes. Other groups would like to use tailocins as an alternative to traditional antibiotics, which indiscriminately eradicate beneficial strains along the bad and are increasingly ineffective due to the development of drug resistance properties.
In their most recent article, the collaborative Berkeley team examined the genetic basis and physical mechanisms that determine how tailocins attack specific strains, and looked at genetic similarities and differences between tailocin producers and their target strains.
After examining 12 strains of soil bacteria known to use tailocins, biologists found evidence that differences in the lipopolysaccharides – fat and sugar molecules – attached to the outer membrane could determine whether a strain was affected by a specific tailocin. targeted or not.
“The bacteria we studied live in a challenging, resource-poor environment, so we’re interested to see how they use tailocins to fight for survival,” said Adam Arkin, co-lead author and senior scientist in the Biosciences, said. Area and technical co-manager of the scientific focus area of the ecosystems and networks integrated with genes and molecular compositions (ENIGMA). Arkin noted that while scientists can easily cause bacteria to produce tailocins in the laboratory (and easily place the genes in culturable strains for mass production, which would come in handy if we want to make tailocins in medicine), there are still many unanswered questions. . about how bacteria tailocins deploy in their natural environment, as well as how – and why – specific strains are targeted with an assassin’s precision.
“Once we understand the targeting mechanisms, we can start using these tailocins ourselves,” Arkin added. “The potential for medicine is huge, of course, but it will also be incredible for the kind of science we do, which studies how environmental microbes interact and the role of these interactions in important ecological processes, such as carbon sequestration and nitrogen processing.”
Currently, it is very difficult to figure out what each microbe does in a community, as scientists cannot easily pick up and subtract strains and observe the outcome. With properly utilized tailocins, these experiments can be easily done.
Mutalik, Arkin and their colleagues are also conducting follow-up studies aimed at revealing the mechanisms of action of tailocins. They plan to use the advanced imaging facilities at Berkeley Lab to take snapshots at the atomic level of the entire process, from the moment the tailocin binds to the target cell to the cell deflation. In essence, they will be filming frames of a microscopic slasher film.
Reference: “Systematic discovery of genetic factors of pseudomonad involved in sensitivity to tailocins” by Sean Carim, Ashley L. Azadeh, Alexey E. Kazakov, Morgan N. Price, Peter J. Walian, Lauren M. Lui, Torben N. Nielsen, Romy Chakraborty, Adam M. Deutschbauer, Vivek K. Mutalik and Adam P. Arkin, March 1, 2021, The ISME Journal.
DOI: 10.1038 / s41396-021-00921-1
This work is part of the ENIGMA Scientific Focus Area, a multi-institutional consortium led by Berkeley Lab that focuses on advancing our understanding of microbial biology and the impact of microbial communities on their ecosystems. ENIGMA is supported by the Department of Energy’s Office of Science.