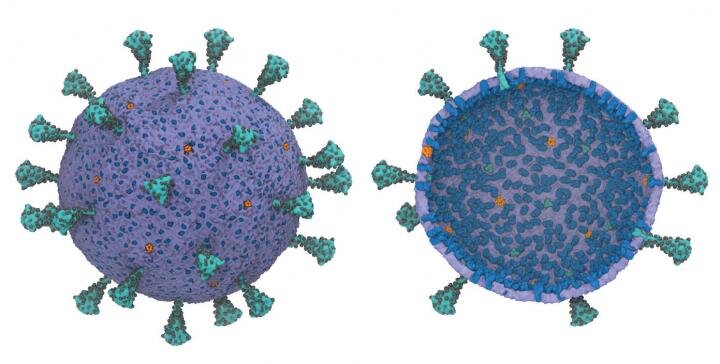
A multi-scale model of the complete SARS-CoV-2 virion was first developed using supercomputers. The model offers scientists the opportunity to exploit new ways of exploiting the vulnerabilities of the virus. Views from outside (L) and inside (R) show the peak protein trimmers (teal), glycosylation sites (black), membrane proteins (blue) and pentameric envelope ion channels (orange). Credit: Gregory Voth, University of Chicago.
The COVID-19 virus contains some mysteries. Scientists remain in the dark about aspects of how it merges and penetrates the host cell; how it gathers itself; and how it emerges from the host cell.
Computational modeling combined with experimental data provides insight into this behavior. But modeling meaningful time scales of the pandemic-causing SARS-CoV-2 virus has so far been limited to just its pieces, such as the ear protein, a target for the current vaccination round.
A new multiscale coarse-grained model of the complete SARS-CoV-2 virion, its nuclear genetic material and virion shell, was developed for the first time using supercomputers. The model offers scientists the opportunity to exploit new ways of exploiting the vulnerabilities of the virus.
“We wanted to understand how SARS-CoV-2 as a whole works holistically,” said Gregory Voth, a professor at the University of Chicago. Voth is the corresponding author of the study that developed the first complete virus model, published in November 2020 in the Biophysical Journal.
“We developed a coarse-grained model from bottom to top,” Voth said, “where we took information from simulations of molecular dynamics at the atomistic level and from experiments.” He explained that a coarse-grained model solves only groups of atoms, as opposed to simulations of all atoms, where each atomic interaction is solved. “If you do it right, which is always a challenge, you maintain the physics in the model.”
The early results of the study show how the vein proteins on the surface of the virus move cooperatively.
“They don’t move independently like a bunch of random, uncorrelated movements,” Voth said. “They work together.”
This cooperative movement of the peak proteins is instructive on how the coronavirus examines and detects the ACE2 receptors of a potential host cell.
“The article we published shows the beginnings of how the motions in the ear proteins correspond,” Voth said. He added that the nails were connected to each other. When one protein moves, another also moves in response.
‘The ultimate aim of the model would be to study, as a first step, the initial virion attractions and interactions with ACE2 receptors on cells and to understand the origin of the attraction and how those proteins work together to continue to deal with the virus fusion process. , ‘Said Voth.
Voth and his group have been developing coarse-grained modeling methods on viruses such as HIV and influenza for more than 20 years. They ‘enlarge’ the data to make it simpler and more computable, while staying true to the dynamics of the system.
“The advantage of the coarse-grained model is that it can be hundreds to thousands of times more effective than the whole-atom model,” Voth explained. The computer savings enabled the team to build a much larger model of the coronavirus than ever before, on longer time scales than was done with all atomic models.
“What you are left with is the much slower, collective movements. The effects of the higher frequency, all atomic movements are folded into the interactions if you do it well. That is the idea of systematic coarse granulation.”
The holistic model developed by Voth began with atomic models of the four main structural elements of the SARS-CoV-2 virion: the vein, membrane, nucleocapsid, and envelope proteins. These atomic models were then simulated and simplified to generate the complete course-shaped model.
The molecular dynamics simulations of all atoms of the vein protein component of the virion system, approximately 1.7 million atoms, were generated by study author Rommie Amaro, a professor of chemistry and biochemistry at the University of California, San Diego.
“Their model basically takes in our data, and it can learn from the data we have on these more detailed scales, and then go beyond where we went,” Amaro said. “This method developed by Voth will enable us and others to simulate scales over the longer period of time needed to simulate the virus that infects a cell.”
Amaro elaborated on the behaviors observed from the coarse-grained simulations of the vein proteins.
“What he saw very clearly was the beginning of the dissociation of the S1 subunit from the peak. The entire upper part of the peak peels off during the merger,” Amaro said.
One of the first steps of viral fusion with the host cell is this dissociation, where it binds to the ACE2 receptor of the host cell.
“The larger S1 aperture motions they saw with this coarse-grained model were something we had not yet seen in the atomic molecular dynamics, and it would be very difficult for us to see,” Amaro said. “It’s a critical part of the function of this protein and the infection process with the host. It was an interesting finding.”
Voth and his team used the dynamic information about the entire atom about the open and closed states of the peak protein generated by the Amaro Lab on the Frontera supercomputer, as well as other data. The Frontera system, funded by the National Science Foundation (NSF), is managed by the Texas Advanced Computing Center (TACC) at the University of Texas at Austin.
“Frontera has shown how important it is for these studies of the virus, on multiple scales. It was critical at the atomic level to understand the underlying dynamics of the peak with all its atoms. There is still much to learn there. But now this information can be used a second time to develop new methods that enable us to go further and further, such as the coarse-grained method, “said Amaro.
“Frontera has been particularly useful in providing the molecular dynamics data at the atomistic level for input into this model. It is very valuable,” Voth said.
The Voth Group initially used the Midway2 computer group at the University of Chicago Research Computing Center to develop the coarse-grained model.
The membrane and sheath protein atom simulations were generated on the Anton 2 system. Anton 2 is operated by the Pittsburgh Supercomputing Center (PSC) with support from the National Institutes of Health and is a special supercomputer for simulations of molecular dynamics developed and presented free of charge by DE Shaw Research.
“Frontera and Anton 2 provided the key input from the molecular level in this model,” Voth said.
“A fantastic thing about Frontera and these kinds of methods is that we can give people much more accurate views on how these viruses move and do their job,” Amaro said.
“There are parts of the virus that are invisible to experiment with,” she continued. “And through these kinds of methods we use on Frontera, our scientists can give the first and most important view of what these systems really look like with all their complexity and how they interact with antibodies or drugs or with parts of the host. cell.”
The type of information Frontera provides to researchers helps to understand the basic mechanisms of viral infection. It is also useful for designing safer and better medicines to treat and prevent the disease, Amaro added.
Voth said: “One thing we are currently concerned about is the UK and the South African SARS-CoV-2 variants. We can probably quickly assess the deviations with a computational platform as we have developed here, what changes are We can hopefully quickly understand the changes these mutations cause to the virus and then hopefully help design new modified vaccines. ”
Scientists create the first computational model of the entire virus responsible for COVID-19
Alvin Yu et al, a coarse-grained model of the SARS-CoV-2 virion, Biophysical Journal (2020). DOI: 10.1016 / j.bpj.2020.10.048
Provided by the University of Texas at Austin
Quotation: First complete coronavirus model shows collaboration (2021, February 26) obtained on February 26, 2021 from https://phys.org/news/2021-02-coronavirus-cooperation.html
This document is subject to copyright. Except for any fair trade for the purpose of private study or research, no portion may be reproduced without the written permission. The content is provided for informational purposes only.