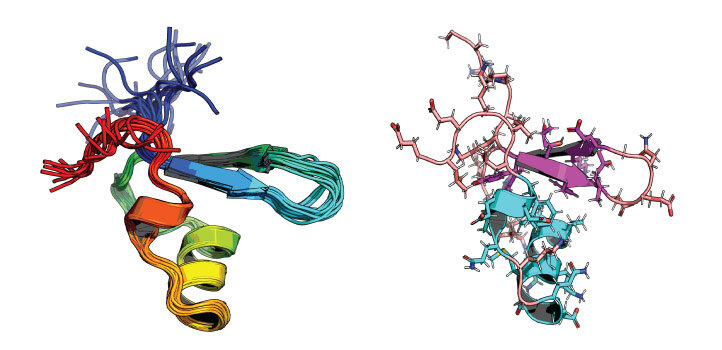
The KAUST team used a synergistic approach to analyze the response of different H-NS proteins (shown above) to temperature and salinity at the atomistic level. Credit: KAUST; Vladlena Kharchenko
Researchers show how bacteria have adapted an observation mechanism that enables them to live in different environments.
A combination of a range of atomic plane techniques has enabled researchers to demonstrate how changes in an environmentally observable protein can survive bacteria in different habitats, from the human gut to deep-sea hydrothermal vents.
“The study gives us an unprecedented insight at the atomic level into how bacteria adapt to changing conditions,” says Stefan Arold, professor of life sciences at KAUST. “To gain these insights, we have shifted the boundaries of three different research methods and put the results together into a unified picture.”
The histone-like nucleoid structuring (H-NS) protein allows bacteria to observe changes in their environment, such as changes in temperature and salinity. Previously, the team showed how the intestinal pathogen uses Salmonella typhimurium H-NS to control its gene expression profile so that it can live optimally inside its warm-blooded host or outside in the soil.
The H-NS protein is also found in bacteria that do not have large temperature fluctuations, such as plant pathogens, insect symbionts and free-living microbes that inhabit hydrothermal vents in the deep sea. Still amazed is how different bacteria adapted the same observation mechanism to suit different lifestyles.
No single analysis technique could unravel the inner workings of this mechanism, and to gain a more integrated view, Arold put together a diverse team of KAUST and international collaborators. Arold and Lukasz Jaremko, a molecular biochemist at KAUST, collaborated with Jianing Li of the University of Vermont to combine several methods: protonless nuclear magnetic resonance spectroscopy, simulations of all atomic molecular dynamics, and biophysical techniques. This synergistic approach enabled the researchers to analyze the reaction of different H-NS proteins at temperature and salinity at the atomistic level.
All H-NS proteins exhibited the same ancestral observation mechanism, whereby temperature and salinity promoted the melting of one of the two dimerization domains of H-NS, releasing the grip on DNA.
However, site-specific amino acid substitutions, mostly in residues involved in salt bridges, have yielded a range of static and dynamic characteristics. These effects dampen or enhance the protein’s response to suit the bacteria’s lifestyle.
“Although the ranges of these proteins are largely conserved, small purposeful changes lead to large differences in their behavior,” says KAUST researcher Umar Farook Shahul Hameed.
Thus, the H-NS protein of the apple pathogen Erwinia amylovoral its sensitivity to heat, which corresponds to the lifestyle of the pathogen in stable temperate climate. And only a few amino acid changes rendered Buchnera’s H-NS aphidicolally the largest environment insensitive, consistent with its role as a mandatory endosimbion of aphids.
“If you tickle the right positions, the behavior changes very easily,” says Arold. “Approaches that interfere with this detection mechanism could find applications in areas ranging from climate change to tackling antibiotic resistance.”
Increases the heat of pathogenic bacteria
Xiaochuan Zhao et al. Molecular basis for the adaptive evolution of environmental observation by H-NS proteins, eLife (2021). DOI: 10.7554 / eLife.57467
eLife
Provided by King Abdullah University of Science and Technology
Quotation: Atomic Techniques Reveals The Evolution Of A Bacterial Protein (2021, March 22) Retrieved March 22, 2021 from https://phys.org/news/2021-03-atomic-techniques-reveal-evolution-bacterial.html
This document is subject to copyright. Apart from any fair trade for the purpose of private study or research, no portion may be reproduced without the written permission. The content is provided for informational purposes only.