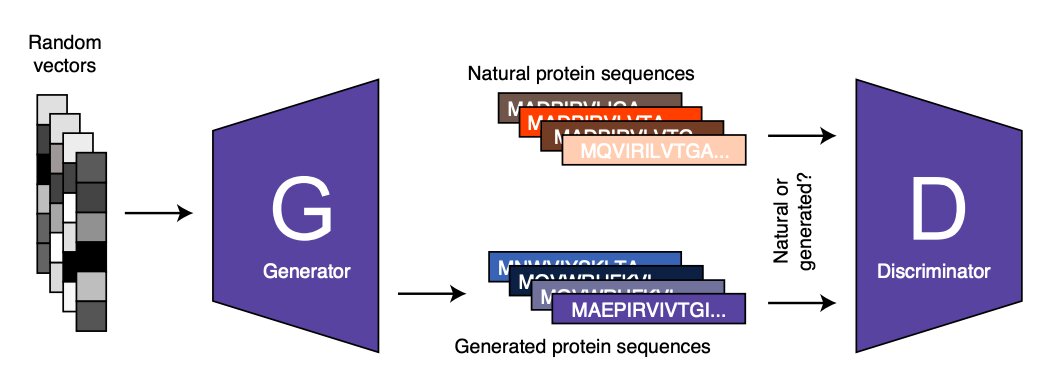
Figure summarizing the training of ProteinGAN. Given a random input vector, the generator network produces a protein sequence that is scored by the discriminator network by comparing it with natural protein sequence. The generator tries to mislead the discriminator by generating sequences that will eventually look like real (the generator never sees real enzyme sequences). Credit: Repecka et al.
Proteins are large, very complex and naturally occurring molecules can be found in all living organisms. These unique substances, which consist of amino acids joined together by peptide bonds to form long chains, can have different functions and properties.
The specific sequence in which different amino acids are arranged to form a given protein ultimately determines the protein’s 3D structure, physicochemical properties, and molecular function. While scientists have been studying proteins for decades, it has so far been very challenging to design proteins that elicit specific chemical reactions.
Researchers at Biomatter Designs, Vilnius University in Lithuania and Chalmers University of Technology in Sweden, recently developed ProteinGAN, a generative adverse network (GAN) that can process and ‘learn’ different natural protein sequences. This unique network, presented in an article published in Natural Machine Intelligence, then uses the information it has obtained to generate new functional protein sequences.
“Proteins are long rows of amino acids that induce processes in all living systems that induce humans,” Alexey Zelezniak, an associate professor at Chalmers University of Technology who led the study, told Phys.org. “Proteins are widely used in our daily lives and are included in countless products, from washing powders to treatments for cancer and coronavirus. It is made up of 20 amino acids arranged in different series and its order determines the function of a protein.”
Creating functional protein sequences is a very challenging task, as even a small change in a given sequence can make a protein non-functional. Non-functional proteins can have harmful and unwanted effects, which can cause humans or animals to develop cancer or other diseases, for example.
“If one wants to make proteins in accordance with human needs, he / she must correctly understand the sequence of amino acids and the given astronomical number of possibilities to make these proteins, which is not a trivial task,” he said. Zelezniak said. “Inspired by the latest developments in AI, especially realistic photo and video generation, we were tempted to ask if current AI technology is ready to produce the most complex molecules humans know proteins.
ProteinGAN, the model developed by Zelezniak and his colleagues, is based on a well-known approach to machine learning, known as adversarial learning. Conflicting learning can be seen as a game that is ‘played’ by two or more artificial neural networks. The first of these networks, known as the ‘generator’, produces a specific type of data (for example an image, text or in the case of ProteinGAN a protein sequence). The second network, known as the ‘discriminator’, tries to distinguish between the artificial data (eg protein sequence) created by the ‘generator’ and authentic or real data.
Next, the generator uses the feedback provided by the discriminator (i.e., the characteristics that made it possible to dispense with generated data apart from actual data) to generate new data. The generator never processes or analyzes real data and the data it produces. Therefore, its learning is based only on the result of the analyzes performed by the discriminator.
“By repeating this process iteratively, both networks get better at what they do, until the generated sequences can not be distinguished from the rights,” Zelezniak said. “Using the AI tool we developed, we were able to generate functional proteins that were active, but that do not yet exist in nature, or that have not yet been discovered.”
In initial trials conducted by the researchers, ProteinGAN generated new and highly diverse protein sequences with physical properties similar to those of natural protein sequences. Using malate dehydrogenase (MDH) as a template enzyme, Zelezniak and his colleagues showed that many of the sequences generated by ProteinGAN are soluble and exhibit MDH catalytic activity, meaning they can have interesting applications in medical and research settings. have. In the future, ProteinGAN could be used to uncover new protein sequences with different properties, which could be valuable for a variety of technological and scientific applications.
“Our research lab focuses on AI-based technologies for synthetic biology applications,” Zelezniak said. “We are currently working to address emerging issues such as plastic pollution, and I believe AI will help build better organisms that are suitable for this particular problem.”
Unique AI method to generate proteins to accelerate drug development
The expansion of functional protein range spaces with generative adversarial networks. Natural Machine Intelligence(2021). DOI: 10.1038 / s42256-021-00310-5.
© 2021 Science X Network
Quotation: ProteinGAN: a generative adversarial network that generates functional protein sequences (2021, April 2) Retrieved April 4, 2021 from https://phys.org/news/2021-04-proteingan-adversarial-network-functional-protein.html
This document is subject to copyright. Except for any fair trade for the purpose of private study or research, no portion may be reproduced without the written permission. The content is provided for informational purposes only.